Tidyverse and read in datas
Day 1
Freie Universität Berlin @ Theoretical Ecology
January 15, 2024
1 The tidyverse

The tidyverse
The tidyverse is an opinonated collection of R packages designed for data science. All packages share an underlying design philosophy, grammar, and data structures.
(www.tidyverse.org)
These are the main packages from the tidyverse that we will use:





Workflow data analysis
The tidyverse
Install the tidyverse once with:
install.packages("tidyverse")Then load and attach the packages at the beginning of your script:
You can also install and load the tidyverse packages individually, but since we will use so many of them together, it’s easier to load and attach them together.
2 Import and export data with readr

Readr
readr is a tidyverse package. To use it, you have to load it:
library(read_csv) The most important functions are:
read_csv/write_csvto read/write comma delimited filesread_tsv/write_tsvto read/write tab delimited filesread_delim/write_delimto read/write files with any delimiter
Read files with read_*()
All read_* functions take a path to the data file as a first argument:
read_*(file = “path/to/your/file”, …)
Import files with a readr function fitting the delimiter of your file:
Use read_delim for a generic type of delimiter:
dat <- read_delim("data/your_data.txt", delim = "\t") # tab delimiter
dat <- read_delim("data/your_data.txt", delim = "..xyz..") # ..xyz.. delimiterAll read_* functions return a tibble
Read files with read_*()
The read functions provide several options to modify the reading of data.
Have a look at ?read_delim for all options.
Useful if your data is not a “perfect table”
Read files with read_*()
Specify number of lines to skip reading with skip
- Useful if you have metadata on top of the file
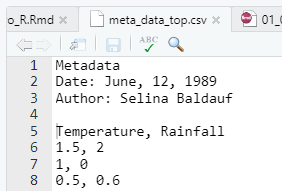
# without skipping first lines
read_csv(file = "data/meta_data_top.csv")# A tibble: 6 × 1
Metadata
<chr>
1 Date: June, 12, 1989
2 Author: Selina Baldauf
3 Temperature, Rainfall
4 1.5, 2
5 1, 0
6 0.5, 0.6 # skip meta data lines
read_csv(
file = "data/meta_data_top.csv",
skip = 4
)# A tibble: 3 × 2
Temperature Rainfall
<dbl> <dbl>
1 1.5 2
2 1 0
3 0.5 0.6Read files with read_*()
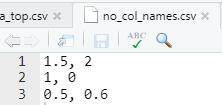
Specify whether the data has a header column or not with col_names
- Useful if you don’t have column names or you want to change them
Read files with read_*()
Specify whether the data has a header column or not with col_names
- Useful if you don’t have column names or you want to change them

# First line expected to be column names
read_csv(file = "data/no_col_names.csv")# A tibble: 2 × 2
`1.5` `2`
<dbl> <dbl>
1 1 0
2 0.5 0.6Write files with write_*()
Every read_* has a corresponding write_* function to export data from R.
Write data from R e.g.
To share transformed or summarized data
Summarize complex raw data and continue working with summarized data
…
Write files with write_*()
All write_* functions take the data to write as the first and the file to write to as the second argument:
write_*(x = dat, file = “path/to/save/file.*”, …)
Use write_delim for a generic type of delimiter:
write_delim(dat, file = "data-clean/your_data.txt", delim = "\t") # tab delimiter
write_delim(dat, file = "data-clean/your_data.txt", delim = "..xyz..") # ..xyz.. delimiter3 Import excel files

Readxl
The readxl package is part of the tidyverse, but you need to load it explicitly
Use the read_excel function to read an excel file:
dat <- read_excel(path = "data/your_data.xlsx")By default, this reads the first sheet. You can read other sheets with
dat <- read_excel(path = "data/your_data.xlsx", sheet = "sheetName") # via sheet name
dat <- read_excel(path = "data/your_data.xlsx", sheet = 2) # via sheet number-
read_excelalso has other functionality, like skipping rows etc. - Check out the package documentation for more functionality
Readxl
A little warning:
- Reading from a text file (.txt or .csv) is more reliable
- Be careful with complicated excel sheets with formulas etc.
- Always double check the data that you imported, e.g. by using the
summaryfunction and checking if the number of rows etc. is correct
Absolute vs. relative paths in R
Absolute paths
C:/Users/Selina/folder1/folder2/data/file_to_read.csv
Relative paths
data/file_to_read.csv
- Relative paths are interpreted relative to the working directory
- Check out where your working directory is with
getwd() - In RStudio projects, the working directory is always the project root
Absolute vs. relative paths
Working with R and RStudio, the best way is to:
-
Organize your work in an RStudio project
- The project root is automatically the working directory
- All your files (also your data) are in one place
- Use paths relative to the project root
Why?
- No need to change the working directory
- Portable paths: will also work on other machines that copied the project
- Makes the code more readable
- Less error prone
4 Guidelines for data sets in
Data format
Follow these guidelines to make data import to R easier and less frustrating
- In general: prefer machine-readable file formats (
.csv,.txtinstead of.xlsx)
Save an Excel spreadsheet as csv
- File -> Save As and select comma separated from the drop down menu
- File -> Export
Data format
Follow these guidelines to make data import to R easier and less frustrating
- In general: prefer machine-readable file formats (
.csv,.txtinstead of.xlsx)
- No white space in column headers
- Use a character as separator, e.g.
species_nameinstead ofspecies name - If this is unpractical, have a look at the function
janitor::clean_names()from thejanitorpackage
- Use a character as separator, e.g.
- No special characters in column headers (ä,, ß, é, ê, %, °C, µ …)
- Use
.as a decimal separator (not,)
Paths and file names
- Avoid white space in paths and file names
-
data-raw/my_data.csvinstead ofdata raw/my data.csv
-
- Avoid special characters in paths
Introduction to R