Data handling with dplyr
Day 2
Freie Universität Berlin @ Theoretical Ecology
January 16, 2024
1 Data transformation
1.1 Data transformation
Data transformation is an important step in understanding the data and preparing it for further analysis.
We can use the tidyverse package dplyr for this.
1.2 Data transformation
With dplyr we can (among other things)
- Filter data to analyse only a part of it
- Create new variables
- Summarize data
- Combine multiple tables
- Rename variables
- Reorder observations or variables
To get started load the package dplyr:
1.3 Dplyr basic vocuabulary for data manipulation
-
filter()picks observations (rows) based on their values -
select()picks variables (columns) based on their names -
mutate()adds new variables based on existing ones -
summarize()combines multiple values into a single summary value
Perform any of these operations by group
1.4 Dplyr basic vocabulary
All of the dplyr functions work similarly:
- First argument is the data (a tibble)
- Other arguments specify what to do exactly
- Return a tibble
1.5 Example data
Soybean production for different use by year and country.
soybean_use <- readr::read_csv('https://raw.githubusercontent.com/rfordatascience/tidytuesday/master/data/2021/2021-04-06/soybean_use.csv')
soybean_use# A tibble: 9,897 × 6
entity code year human_food animal_feed processed
<chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 Africa <NA> 1961 33000 6000 14000
2 Africa <NA> 1962 43000 7000 17000
3 Africa <NA> 1963 31000 7000 5000
4 Africa <NA> 1964 43000 6000 14000
5 Africa <NA> 1965 34000 6000 12000
6 Africa <NA> 1966 41000 6000 2000
7 Africa <NA> 1967 47000 6000 4000
8 Africa <NA> 1968 50000 7000 3000
9 Africa <NA> 1969 52000 6000 6000
10 Africa <NA> 1970 52000 6000 8000
# ℹ 9,887 more rows
2 filter()
picks observations (rows) based on their value

Artwork by Allison Horst
2.1 Useful filter() helpers
These functions and operators help you filter your observations:
2.2 filter()
Filter rows that contain the values for Germany
filter(soybean_use, entity == "Germany")# A tibble: 53 × 6
entity code year human_food animal_feed processed
<chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 Germany DEU 1961 0 3000 1042000
2 Germany DEU 1962 0 3000 935000
3 Germany DEU 1963 0 3000 1092000
4 Germany DEU 1964 0 3000 1096000
5 Germany DEU 1965 0 3000 1435000
6 Germany DEU 1966 0 3000 1588000
7 Germany DEU 1967 0 3000 1646000
8 Germany DEU 1968 0 3000 1480000
9 Germany DEU 1969 0 3000 1423000
10 Germany DEU 1970 0 3000 2118000
# ℹ 43 more rowsfilter() goes through each row of the data and return only those rows where the value for entity is "Germany"
2.3 filter() + %in%
Use the %in% operator to filter rows based on multiple values, e.g. countries
countries_select <- c("Germany", "Austria", "Switzerland")
filter(soybean_use, entity %in% countries_select)# A tibble: 159 × 6
entity code year human_food animal_feed processed
<chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 Austria AUT 1961 0 0 0
2 Austria AUT 1962 0 0 0
3 Austria AUT 1963 0 0 0
4 Austria AUT 1964 0 0 0
5 Austria AUT 1965 0 0 0
6 Austria AUT 1966 0 0 0
7 Austria AUT 1967 0 0 0
8 Austria AUT 1968 0 0 0
9 Austria AUT 1969 0 0 0
10 Austria AUT 1970 0 0 0
# ℹ 149 more rows
2.4 filter() + is.na()
Filter only rows that don’t have a country code (i.e. the continents etc.)
# A tibble: 1,734 × 6
entity code year human_food animal_feed processed
<chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 Africa <NA> 1961 33000 6000 14000
2 Africa <NA> 1962 43000 7000 17000
3 Africa <NA> 1963 31000 7000 5000
4 Africa <NA> 1964 43000 6000 14000
5 Africa <NA> 1965 34000 6000 12000
6 Africa <NA> 1966 41000 6000 2000
7 Africa <NA> 1967 47000 6000 4000
8 Africa <NA> 1968 50000 7000 3000
9 Africa <NA> 1969 52000 6000 6000
10 Africa <NA> 1970 52000 6000 8000
# ℹ 1,724 more rows
2.5 filter() + between()
Combine different filters:
Select rows where
- the value for
yearsis between 1970 and 1980 - the value for
entityis Germany
# A tibble: 11 × 6
entity code year human_food animal_feed processed
<chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 Germany DEU 1970 0 3000 2118000
2 Germany DEU 1971 0 3000 2119000
3 Germany DEU 1972 0 5000 2271000
4 Germany DEU 1973 0 3000 2820000
5 Germany DEU 1974 0 3000 3704000
6 Germany DEU 1975 0 3000 3480000
7 Germany DEU 1976 0 1000 3453000
8 Germany DEU 1977 0 3000 3388000
9 Germany DEU 1978 0 3000 3647000
10 Germany DEU 1979 0 2000 3700000
# ℹ 1 more row
3 select()
picks variables (columns) based on their names
3.1 Useful select() helpers
-
starts_with()andends_with(): variable names that start/end with a specific string -
contains(): variable names that contain a specific string -
matches(): variable names that match a regular expression -
any_of()andall_of(): variables that are contained in a character vector
3.2 select()
Select the variables entity, year and human food
select(soybean_use, entity, year, human_food)# A tibble: 9,897 × 3
entity year human_food
<chr> <dbl> <dbl>
1 Africa 1961 33000
2 Africa 1962 43000
3 Africa 1963 31000
4 Africa 1964 43000
5 Africa 1965 34000
6 Africa 1966 41000
7 Africa 1967 47000
8 Africa 1968 50000
9 Africa 1969 52000
10 Africa 1970 52000
# ℹ 9,887 more rowsRemove variables using -
select(soybean_use, -entity, -year, -human_food)
3.3 select() + ends_with()
Select all columns that end with "d"
# A tibble: 9,897 × 3
human_food animal_feed processed
<dbl> <dbl> <dbl>
1 33000 6000 14000
2 43000 7000 17000
3 31000 7000 5000
# ℹ 9,894 more rowsYou can use the same structure for starts_with() and contains().
# this does not match any rows in the soy bean data set
# but combinations like this are helpful for research data
select(soybean_use, starts_with("sample_"))
select(soybean_use, contains("_id_"))
3.4 select() + any_of()/all_of()
Use a character vector in conjunction with column selection
cols <- c("sample_", "year", "processed", "entity")any_of() returns any columns that match an element in cols
# A tibble: 9,897 × 3
year processed entity
<dbl> <dbl> <chr>
1 1961 14000 Africa
# ℹ 9,896 more rows
3.5 select() + from:to
Multiple consecutive columns can be selected using the from:to structure with either column id or variable name:
# A tibble: 9,897 × 4
code year human_food animal_feed
<chr> <dbl> <dbl> <dbl>
1 <NA> 1961 33000 6000
2 <NA> 1962 43000 7000
3 <NA> 1963 31000 7000
# ℹ 9,894 more rowsBe a bit careful with these commands: They are not robust if you e.g. change the order of your columns at some point.
4 mutate()
Adds new variables

Artwork by Allison Horst
4.1 mutate()
New columns can be added based on values from other columns
mutate(soybean_use,
sum_human_animal = human_food + animal_feed
)# A tibble: 9,897 × 7
entity code year human_food animal_feed processed sum_human_animal
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Africa <NA> 1961 33000 6000 14000 39000
2 Africa <NA> 1962 43000 7000 17000 50000
3 Africa <NA> 1963 31000 7000 5000 38000
# ℹ 9,894 more rowsAdd multiple new columns at once:
mutate(soybean_use,
sum_human_animal = human_food + animal_feed,
total = human_food + animal_feed + processed
)
4.2 mutate() + case_when()
Use case_when to add column values conditional on other columns.
case_when() can combine many cases into one.
mutate(soybean_use,
legislation = case_when(
between(year, 1980, 2000) ~ "legislation_1", # case 1
year >= 2000 ~ "legislation_2", # case 2
.default = "no_legislation" # all other cases
)
)# A tibble: 9,897 × 7
entity code year human_food animal_feed processed legislation
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <chr>
1 Africa <NA> 1961 33000 6000 14000 no_legislation
2 Africa <NA> 1962 43000 7000 17000 no_legislation
3 Africa <NA> 1963 31000 7000 5000 no_legislation
4 Africa <NA> 1964 43000 6000 14000 no_legislation
5 Africa <NA> 1965 34000 6000 12000 no_legislation
6 Africa <NA> 1966 41000 6000 2000 no_legislation
7 Africa <NA> 1967 47000 6000 4000 no_legislation
8 Africa <NA> 1968 50000 7000 3000 no_legislation
9 Africa <NA> 1969 52000 6000 6000 no_legislation
10 Africa <NA> 1970 52000 6000 8000 no_legislation
# ℹ 9,887 more rows
5 summarize()
summarizes data
5.1 summarize()
summarize will collapse the data to a single row
5.2 summarize() by group
summarize is much more useful in combination with the grouping argument .by
- summary will be calculated separately for each group
# summarize the grouped data
summarize(soybean_use,
total_animal = sum(animal_feed, na.rm = TRUE),
total_human = sum(human_food, na.rm = TRUE),
.by = year
)# A tibble: 53 × 3
year total_animal total_human
<dbl> <dbl> <dbl>
1 1961 1503000 16994000
2 1962 1800000 17326000
3 1963 2060000 18667000
4 1964 2002000 19639000
5 1965 2162000 17796000
6 1966 3096000 22179000
7 1967 2818000 23282000
8 1968 3361000 22747000
9 1969 3084000 22212000
10 1970 2496000 24119000
# ℹ 43 more rows
5.3 count()
Counts observations by group
# count rows grouped by year
count(soybean_use, year)# A tibble: 53 × 2
year n
<dbl> <int>
1 1961 178
2 1962 178
3 1963 178
4 1964 178
5 1965 178
6 1966 178
7 1967 178
8 1968 178
9 1969 178
10 1970 178
# ℹ 43 more rows
6 The pipe %>%
Combine multiple data operations into one command
6.1 The pipe %>%
Data transformation often requires multiple operations in sequence.
The pipe operator %>% helps to keep these operations clear and readable.
- You may also see
|>from the base R
6.2 The pipe %>%
Let’s look at an example without pipe:
How could we make this more efficient?
6.3 The pipe %>%
We could do everything in one step without intermediate results by using use one nested function
But this gets complicated and error prone very quickly
6.4 The pipe %>%
The pipe operator makes it very easy to combine multiple operations:
You can read from top to bottom and interpret the %>% as an “and then do”.
6.5 The pipe %>%
But what is happening?
The pipe is “pushing” the result of one line into the first argument of the function from the next line.
Piping works perfectly with the tidyverse functions because they are designed to return a tibble and take a tibble as first argument.
Tip
Use the keyboard shortcut Ctrl/Cmd + Shift + M to insert %>%
6.6 The pipe %>%
Piping also works well together with ggplot
7 Combining mulitiple tables
7.1 Combine two tibbles by row bind_rows
Situation: Two (or more) tibbles with the same variables (column names)
tbl_a <- soybean_use[1:2, ] # first two rows
tbl_b <- soybean_use[2:nrow(soybean_use), ] # the resttbl_a# A tibble: 2 × 6
entity code year human_food animal_feed processed
<chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 Africa <NA> 1961 33000 6000 14000
2 Africa <NA> 1962 43000 7000 17000tbl_b# A tibble: 9,896 × 6
entity code year human_food animal_feed processed
<chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 Africa <NA> 1962 43000 7000 17000
2 Africa <NA> 1963 31000 7000 5000
# ℹ 9,894 more rows
7.2 Combine two tibbles by row bind_rows
Bind the rows together with bind_rows():
bind_rows(tbl_a, tbl_b)# A tibble: 9,898 × 6
entity code year human_food animal_feed processed
<chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 Africa <NA> 1961 33000 6000 14000
2 Africa <NA> 1962 43000 7000 17000
# ℹ 9,896 more rowsYou can also add an ID-column to indicate which line belonged to which table:
bind_rows(a = tbl_a, b = tbl_b, .id = "id")# A tibble: 9,898 × 7
id entity code year human_food animal_feed processed
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 a Africa <NA> 1961 33000 6000 14000
2 a Africa <NA> 1962 43000 7000 17000
3 b Africa <NA> 1962 43000 7000 17000
# ℹ 9,895 more rowsYou can use bind_rows() to bind as many tables as you want:
bind_rows(a = tbl_a, b= tbl_b, c = tbl_c, ..., .id = "id")
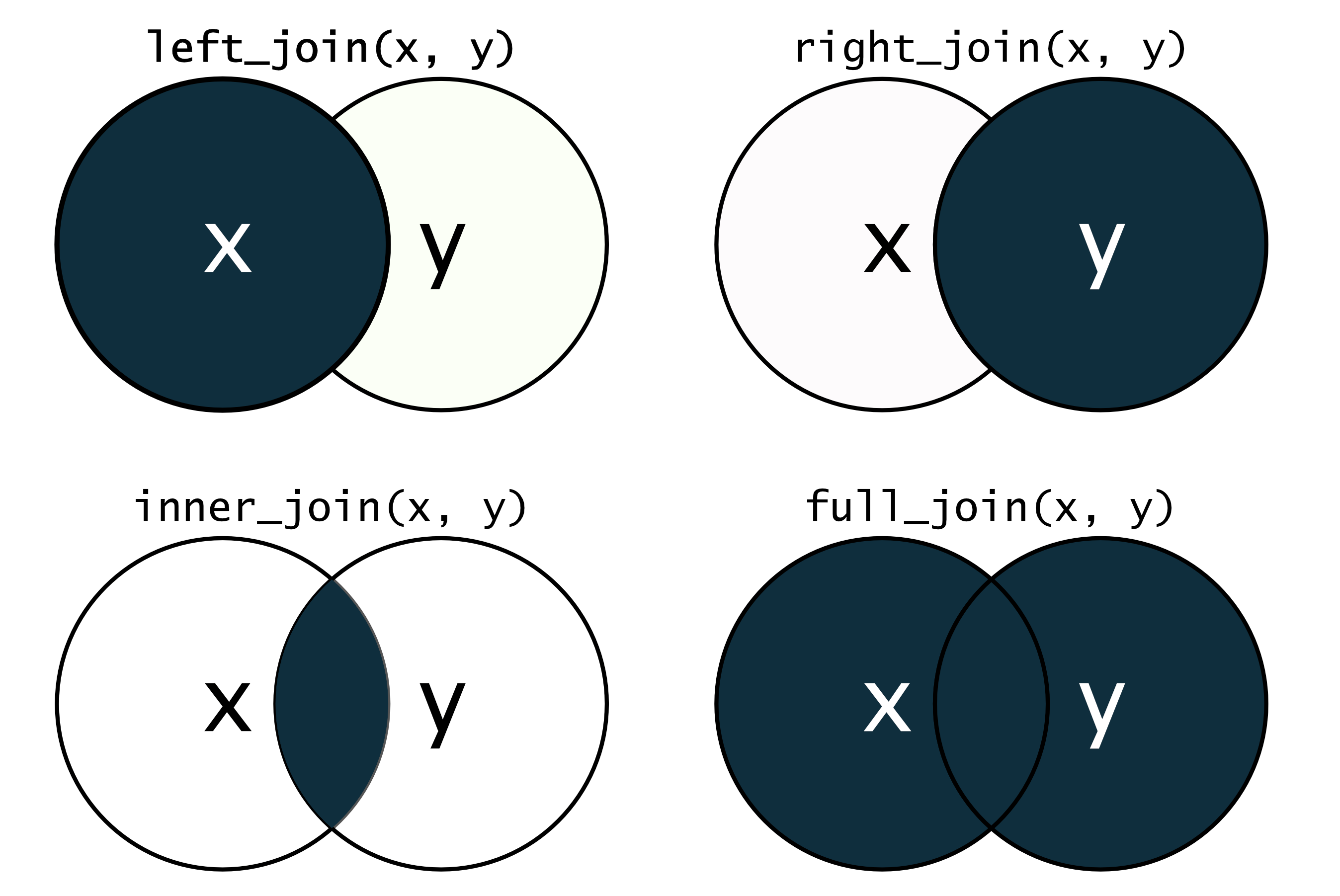
7.3 Join tibbles with left_join()
Situation: Two tables that share some but not all columns.
soybean_use# A tibble: 9,897 × 6
entity code year human_food animal_feed processed
<chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 Africa <NA> 1961 33000 6000 14000
2 Africa <NA> 1962 43000 7000 17000
# ℹ 9,895 more rows# table with the gdp of the country/continent for each year
gdp# A tibble: 9,897 × 3
entity year gdp
<chr> <dbl> <dbl>
1 Africa 1961 5.79
2 Africa 1962 5.79
# ℹ 9,895 more rows
7.4 Join tibbles with left_join()
Join the two tables by the two common columns entity and year
# A tibble: 9,897 × 7
entity code year human_food animal_feed processed gdp
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Africa <NA> 1961 33000 6000 14000 5.79
2 Africa <NA> 1962 43000 7000 17000 5.79
3 Africa <NA> 1963 31000 7000 5000 5.79
4 Africa <NA> 1964 43000 6000 14000 5.79
5 Africa <NA> 1965 34000 6000 12000 5.79
6 Africa <NA> 1966 41000 6000 2000 5.79
7 Africa <NA> 1967 47000 6000 4000 5.79
8 Africa <NA> 1968 50000 7000 3000 5.79
9 Africa <NA> 1969 52000 6000 6000 5.79
10 Africa <NA> 1970 52000 6000 8000 5.79
# ℹ 9,887 more rowsleft_join() means that the resulting tibble will contain all rows of soybean_use, but not necessarily all rows of gdp
7.5 Different *_join() functions
8 Summary
Data transformation with dplyr
8.1 Summary I
All dplyr functions take a tibble as first argument and return a tibble.
filter()
8.2 Summary II
All dplyr functions take a tibble as first argument and return a tibble.
select()
- pick columns with helpers
8.3 Summary III
arrange()
-
change order of rows (adscending)
- or descending with
desc()
- or descending with
mutate()
-
add columns but keep all columns
-
case_when()for conditional values
-
8.4 Summary IV
summarize()
-
collapse rows into one row by some summary
- use
.byargument to summarize by group
- use
count
- count rows based on a group
8.5 Summary V
bind_rows()
-
combine rows of multiple tibbles into one
- the tibbles need to have the same columns
- add an id column with the argument
.id = "id" - function
bind_cols()works similarly just for columns
left_join()
- combine tables based on common columns
Introduction to R