library(dplyr)
library(tidyr)
library(ggplot2)
## theme for ggplot
theme_set(theme_classic())
theme_update(text = element_text(size = 14))Statistical tests
Day 5
Freie Universität Berlin @ Theoretical Ecology
January 19, 2024
Reproduce slides
1 Introduction to null-hypothesis significance testing
procedure:
- formulate a null hypothesis (\(H_0\)) and an alternative hypothesis (\(H_1\))
- collect data
- calculate the probability of observing data at least as extreme if \(H_0\) was true (p-value)
- if p-value < \(\alpha\): reject \(H_0\) (typically \(\alpha = 0.05\))
2 P-values
- is used in null-hypothesis significance testing
- is the probability of obtaining test results at least as extreme as the result actually observed, under the assumption that the null hypothesis is correct
Which factors influence the p-value?
- Variability in the data
- Effect size
- Sample size
- please report all of them in your report/article
3 Simulation study on p-values
Life expectancy
- simulate life expectancy of 100 people with a log-normal distribution
- we then compare the life expectancy of two groups (control, treatment) with a Wilcoxon test
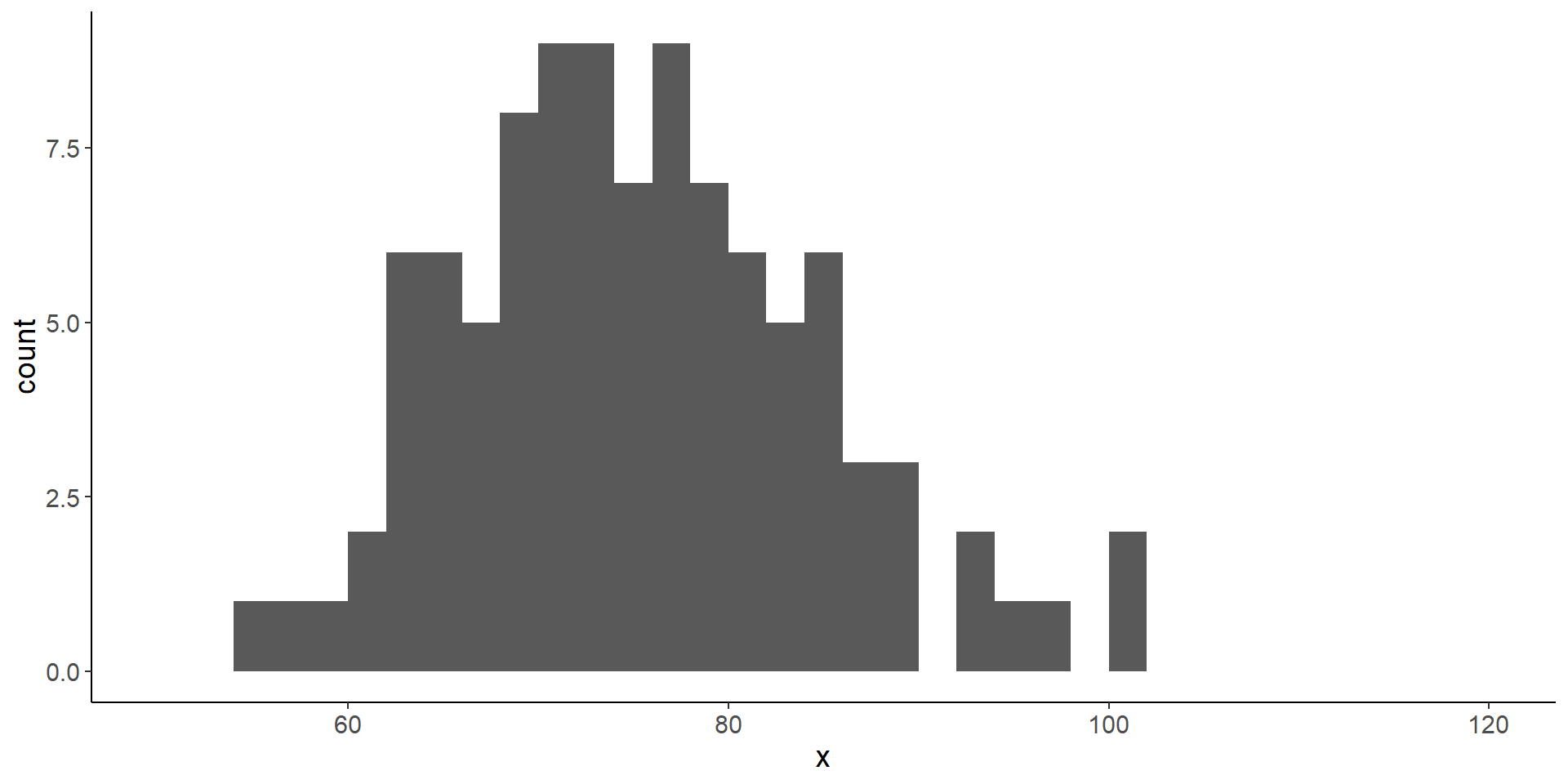
No difference in life expectancy
p_values <- c()
for (i in 1:2000) {
x_control <- rlnorm(100, meanlog = log(75), sdlog = 0.12)
x_treatment <- rlnorm(100, meanlog = log(75), sdlog = 0.12)
p_val <- wilcox.test(x_control, x_treatment)$p.value
p_values <- c(p_values, p_val)
}
mean(p_values < 0.05)[1] 0.051Code
plotting_df <- tibble(p_values = p_values,
color = ifelse(p_values < 0.05, "significant", "non-significant"))
ggplot(plotting_df) +
geom_histogram(aes(p_values, fill = color), bins = 100,
breaks = seq(0, 1, 0.05)) +
scale_fill_manual(values = c("significant" = "red", "non-significant" = "grey")) +
labs(fill = "", x = "p-value")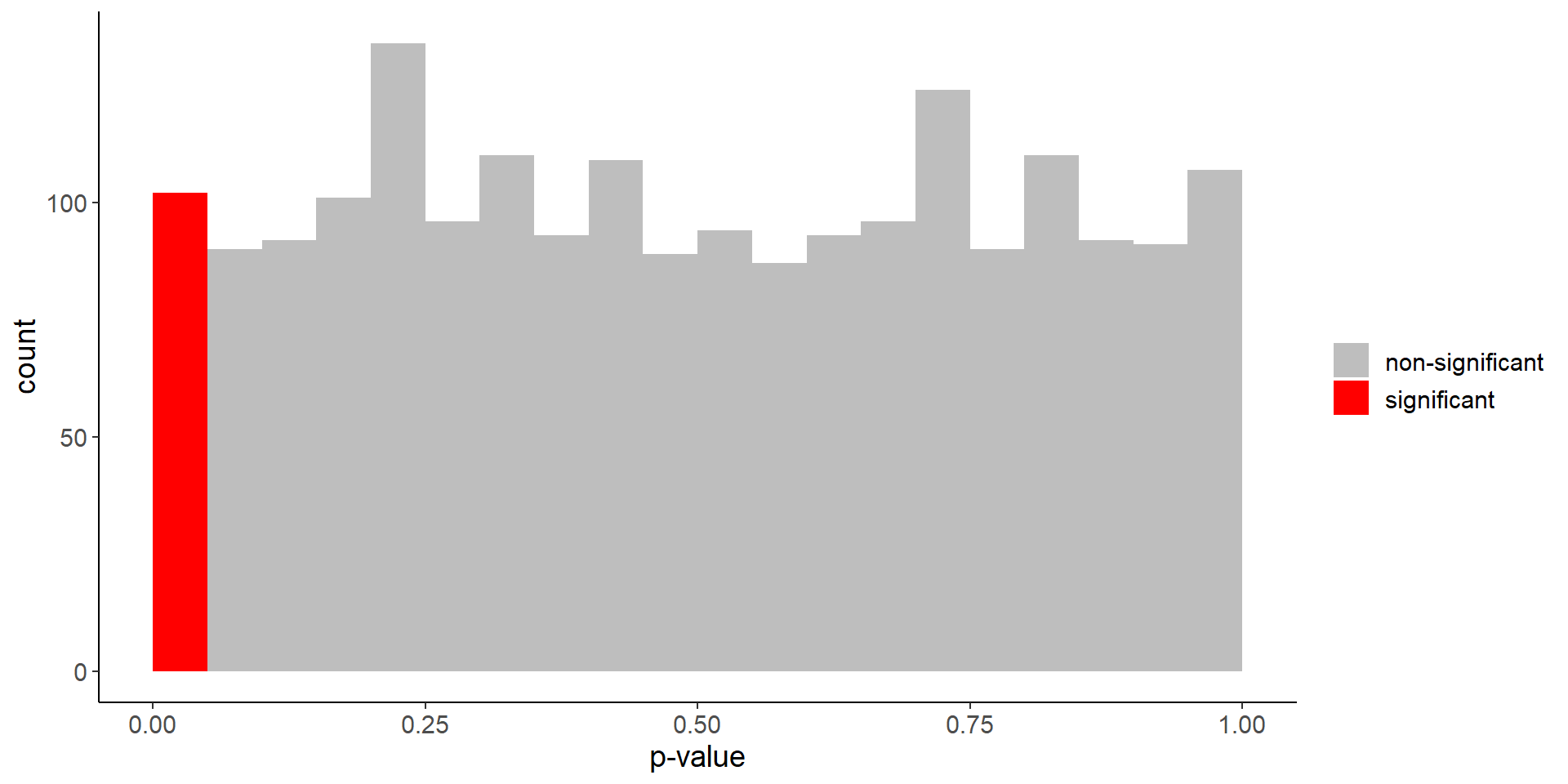
Difference in life expectancy
p_values <- c()
for (i in 1:2000) {
x_control <- rlnorm(100, meanlog = log(75), sdlog = 0.12)
x_treatment <- rlnorm(100, meanlog = log(77), sdlog = 0.12)
p_val <- wilcox.test(x_control, x_treatment)$p.value
p_values <- c(p_values, p_val)
}
mean(p_values < 0.05)[1] 0.317Code
plotting_df <- tibble(p_values = p_values,
color = ifelse(p_values < 0.05, "significant", "non-significant"))
ggplot(plotting_df) +
geom_histogram(aes(p_values, fill = color), bins = 100,
breaks = seq(0, 1, 0.05)) +
scale_fill_manual(values = c("significant" = "red", "non-significant" = "grey")) +
labs(fill = "", x = "p-value")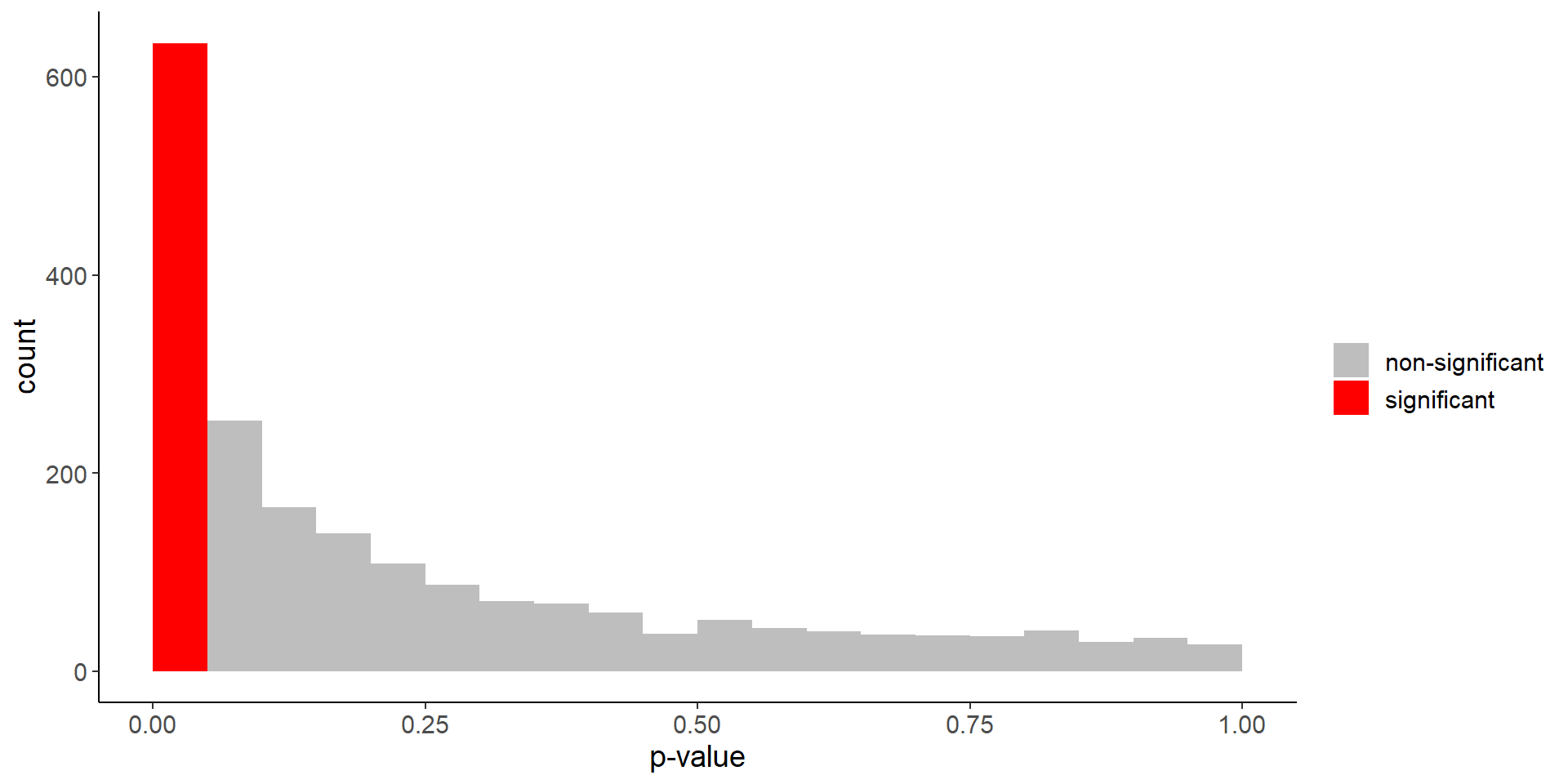
Difference in life expectancy
p_values <- c()
for (i in 1:2000) {
x_control <- rlnorm(10, meanlog = log(75), sdlog = 0.12)
x_treatment <- rlnorm(10, meanlog = log(77), sdlog = 0.12)
p_val <- wilcox.test(x_control, x_treatment)$p.value
p_values <- c(p_values, p_val)
}
mean(p_values < 0.05)[1] 0.061Code
plotting_df <- tibble(p_values = p_values,
color = ifelse(p_values < 0.05, "significant", "non-significant"))
ggplot(plotting_df) +
geom_histogram(aes(p_values, fill = color), bins = 100,
breaks = seq(0, 1, 0.05)) +
scale_fill_manual(values = c("significant" = "red", "non-significant" = "grey")) +
labs(fill = "", x = "p-value")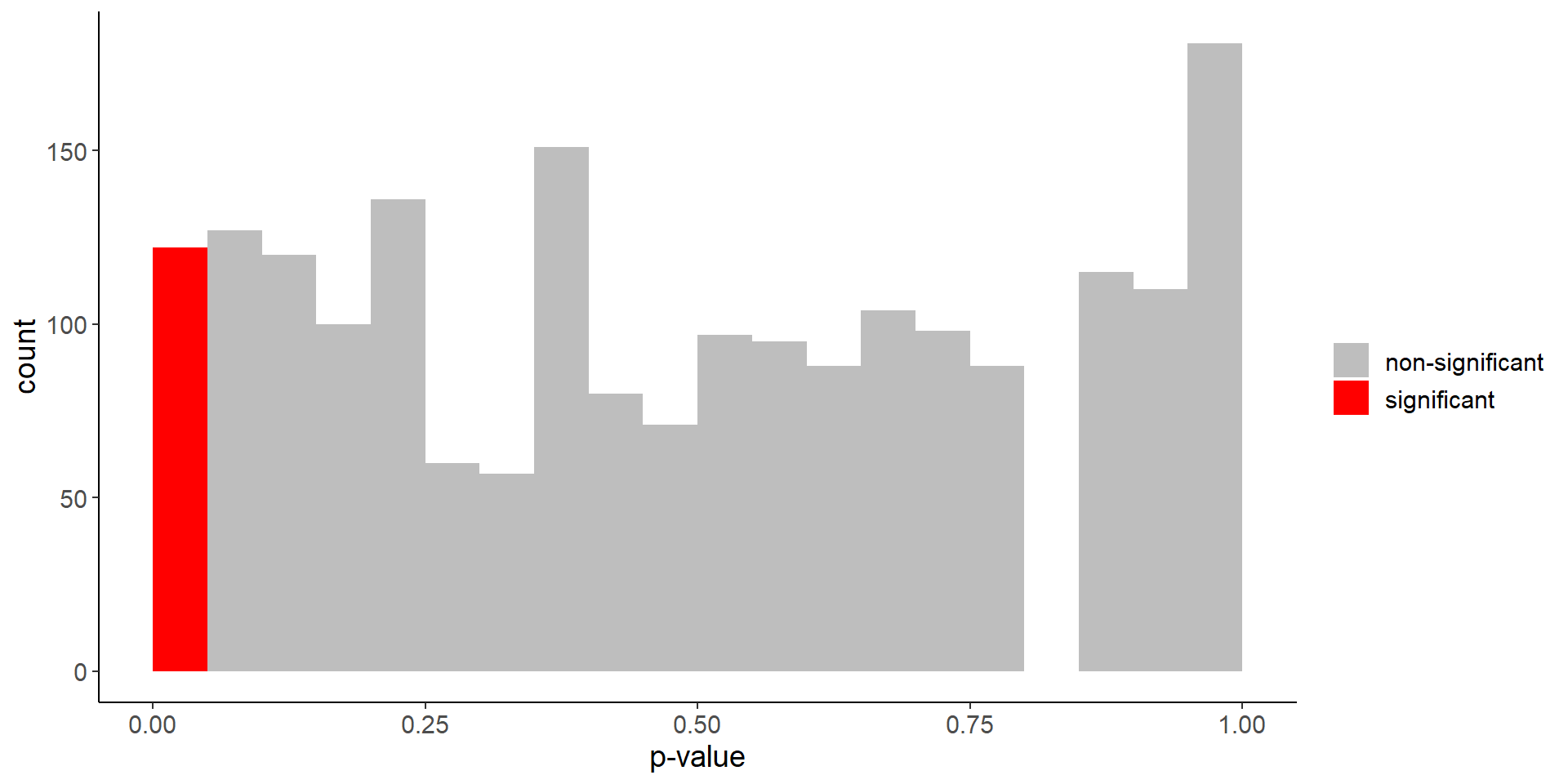
Difference in life expectancy
p_values <- c()
for (i in 1:2000) {
x_control <- rlnorm(100, meanlog = log(75), sdlog = 0.12)
x_treatment <- rlnorm(100, meanlog = log(80), sdlog = 0.12)
p_val <- wilcox.test(x_control, x_treatment)$p.value
p_values <- c(p_values, p_val)
}
mean(p_values < 0.05)[1] 0.9595Code
plotting_df <- tibble(p_values = p_values,
color = ifelse(p_values < 0.05, "significant", "non-significant"))
ggplot(plotting_df) +
geom_histogram(aes(p_values, fill = color), bins = 100,
breaks = seq(0, 1, 0.05)) +
scale_fill_manual(values = c("significant" = "red", "non-significant" = "grey")) +
labs(fill = "", x = "p-value")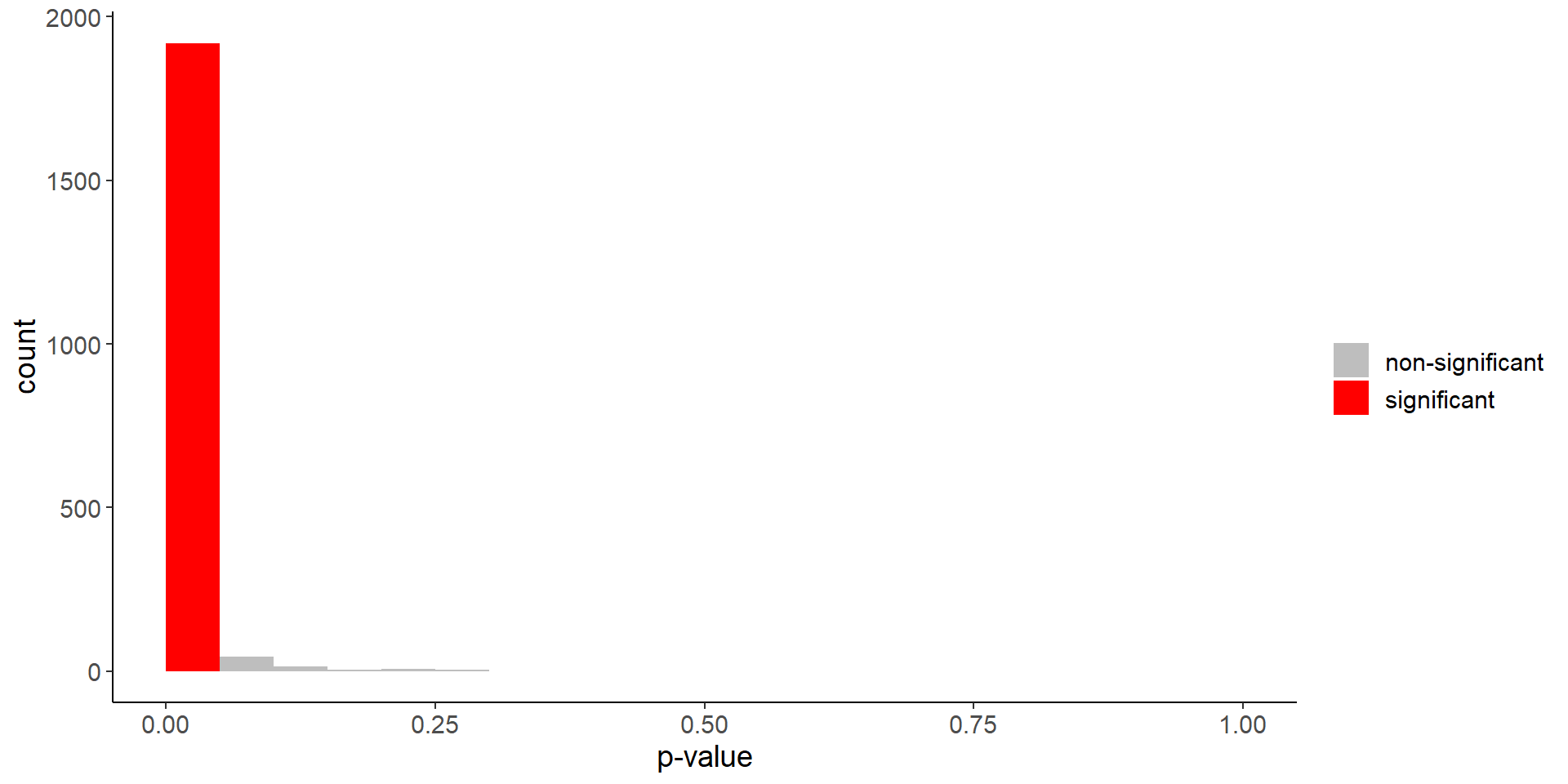
4 Errors in statistical tests
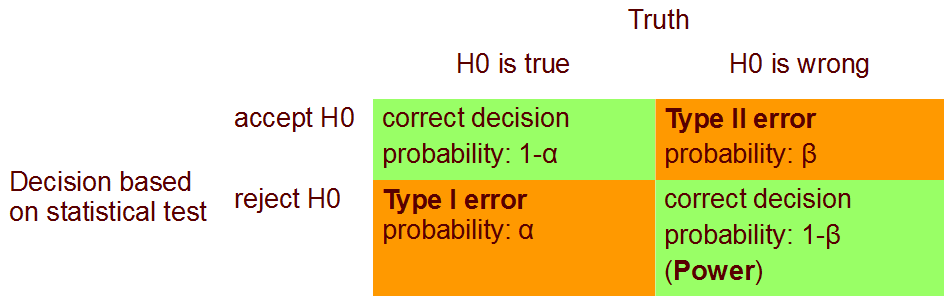
5 Pitfalls - thinks to keep in mind
- statistical significance does not imply biological significance
- the null hypothesis is almost never true
- no statistical significance does not imply no effect
- arbitrary choice of \(\alpha\) (typically 0.05)
see e.g. here
Please dont’t do:
- p-hacking: trying out different tests until you find a significant result
- data dredging: trying out different subsets of your data until you find a significant result
- multiple testing: doing multiple tests on the same data until you find a significant result (without correcting for multiple testing)
6 Test for equal means (distribution)
t-test
- Normal distribution AND equal variance
- Compares if mean values are within range of standard error of each other
- p: how likely is the difference if the means were equal
Welch-Test
- Normal distribution but unequal variance
- Corrected t-test
Wilcoxon rank sum test (Mann–Whitney U test)
- Non-normal distribution and unequal variance
- Compares rank sums of the data
- Non-parametric
Example dataset
InsectSprays: counts of insects in agricultural experimental units treated with different insecticides
t-test
\(H_0\): The samples do not differ in their mean
Treatment A and B: normally distributed and equal variance
t.test(TreatA, TreatB, var.equal = TRUE)
Two Sample t-test
data: TreatA and TreatB
t = -0.45352, df = 22, p-value = 0.6546
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-4.643994 2.977327
sample estimates:
mean of x mean of y
14.50000 15.33333 - t: test statistics (t = 0 means equal means)
- df: degrees of freedom of t-statistics
- p-value: how likely is it to observe the data if \(H_0\) was true?
t-test
\(H_0\): The samples do not differ in their mean
Treatment A and B: normally distributed and equal variance
t.test(TreatA, TreatB, var.equal = TRUE)
Two Sample t-test
data: TreatA and TreatB
t = -0.45352, df = 22, p-value = 0.6546
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-4.643994 2.977327
sample estimates:
mean of x mean of y
14.50000 15.33333 Result: The means of spray A and B don’t differ significantly (t = -0.45, df = 22, p = 0.66)
Welch-Test
\(H_0\): The samples do not differ in their mean
Treatment A and C: normally distributed and non-equal variance
t.test(TreatA, TreatC, var.equal = FALSE)
Welch Two Sample t-test
data: TreatA and TreatC
t = 8.4073, df = 14.739, p-value = 5.278e-07
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
9.263901 15.569433
sample estimates:
mean of x mean of y
14.500000 2.083333 Result: The means of spray A and C do differ significantly (t = 7.58, df = 13.9, p < 0.001)
Wilcoxon-rank-sum Test (Mann–Whitney U test)
\(H_0\): For randomly selected values X and Y from two populations, the probability of X being greater than Y is equal to the probability of Y being greater than X (if both populations have a similar type of distribution and variance: both medians are equal).
wilcox.test(TreatA, TreatB)
Wilcoxon rank sum test with continuity correction
data: TreatA and TreatB
W = 62, p-value = 0.5812
alternative hypothesis: true location shift is not equal to 0Result: We cannot reject the null hypothesis that the probability of counted insects from treatment A being greater than the counted insects from treatment B is equal to the probability of the counted insects from treatment B being greater than the counted insects from treatment A (W = 62, p = 0.58).
Paired values
Are there pairs of data points?
Example: samples of invertebrates across various rivers before and after sewage plants.
- For each plant, there is a pair of data points (before and after the plant)
- Question: Is the change (before-after) significant
Use paired = TRUE in the test.
t.test(TreatA, TreatB, var.equal = TRUE, paired = TRUE)
t.test(TreatA, TreatB, var.equal = FALSE, paired = TRUE)
wilcox.test(TreatA, TreatB, paired = TRUE)Careful: your treatment vector both have to have the same order
Note: Plot mean +- se using stat_summary
One way to plot the results is to plot mean and standard error of the mean:
ggplot(InsectSprays, aes(x = spray, y = count)) +
geom_jitter(width = 0.2, height = 0, alpha = 0.2) +
stat_summary(fun.data = mean_se)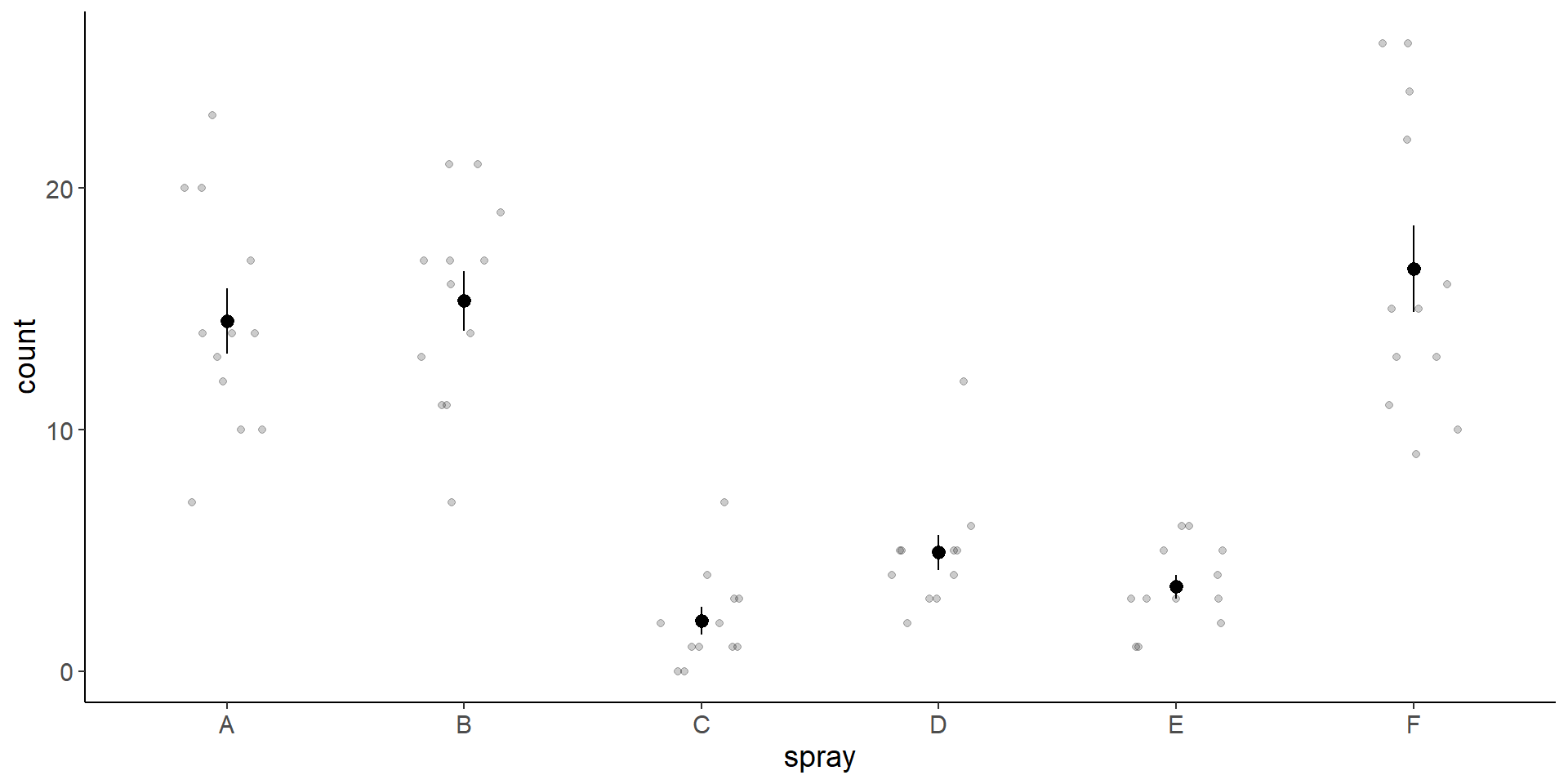
Statistical tests