library(tidyr)
library(dplyr)
library(ggplot2)
library(readr)
library(forcats)
## theme for ggplot
theme_set(theme_classic())
theme_update(text = element_text(size = 14))Overdispersion in GLMs
Day 8
Felix May
Freie Universität Berlin @ Theoretical Ecology
Reproduce slides
Overdispersion
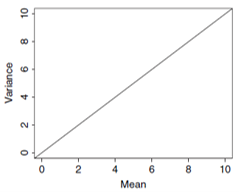
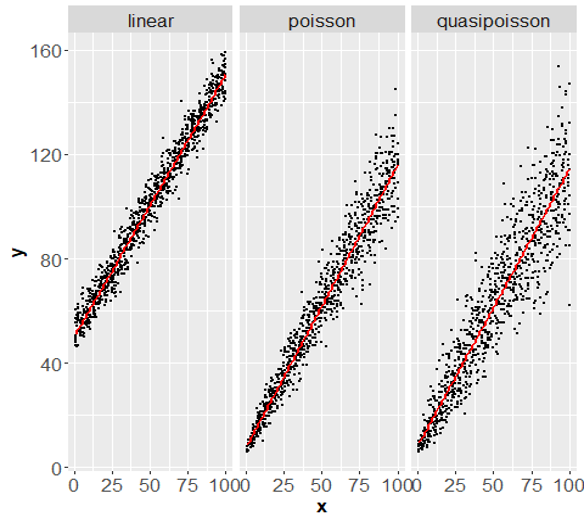
- Poisson GLM
\(\text{mean}(y_i) = \lambda_i\)
\(\text{var}(y_i) = \lambda_i\)
\(\text{var}(y_i) > \lambda_i\)
- Binomial GLM
\(\text{mean}(y_i) = N_i p_i\)
\(\text{var}(y_i) = N_i p_i (1 - p_i)\)
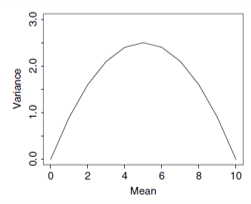
\(\text{var}(y_i) > N_i p_i (1 - p_i)\)
Overdispersion: Higher variance than expected from error distribution
Consequences of overdispersion
Ignoring overdispersion
- Overestimation of significance, i.e. p-values biased to lower values
- Inappropriate hypotheses tests
- Biased predictions
- Check for overdispersion in Poisson and Binomial-GLMs
- Not needed for binary data!
Detecting overdispersion
- Ratio: residual deviance / residual degrees of freedom (df)
- Residual deviance / df > 1.5 –> overdispersion
- Residual deviance / df < 0.6 –> underdispersion (rare in the real world)
Model overdispersion
- Two approaches
- Quasi-Likelihood approach
- Count data –> quasipoisson
- Proportion data –> quasibinomial
- Different error distribution
- Count data: Negative binomial distribution
- Extra parameter for the increase in variance relative to Poisson distribution
- Count data: Negative binomial distribution
Quasi-Likelihood approach
- Assumption: Proportional relationship between \(\text{mean}(y_i)\) and \(\text{var}(y_i)\)
- Poisson: \(\text{var}(y_i) = \phi \lambda_i\)
- Binomial: \(\text{var}(y_i) = \phi N_i p_i (1-p_i)\)
- Estimate \(\phi\) from data \(\phi \approx \frac{\text{Residual deviance}}{\text{df}}\)
- Consequences
- Equal parameter estimates than with \(\phi = 1\)
- Inflation of standard erros by \(\sqrt{\phi}\)
Quasi-Likelihood approach
- Linear regression
- \(\sigma^2 = c\)
- Poisson GLM
- \(\sigma^2 = \lambda\)
- Quasipoisson GLM
- \(\sigma^2 = \phi\lambda\)
Negative binomial distribution
- Discrete distribution for count data
- Relation to Binomial dist.: No. of failures, until you find a given no. of successes
- Relation to Poisson dist.: Poisson distribution with variance in \(\lambda\)
- Different parametrizations in the literature
- \(\mu\) … mean
- \(\theta\) .. dispersion parameter
- Large –> low overdispersion
- Small –> large overdispersion
\(\text{mean}(y_i) = \mu_i\)
\(\text{var}(y_i) = \mu_i + \frac{\mu_i^2}{\theta}\)
Negative binomial distribution
Code
x <- 0:25
ypois1 <- dpois(x, lambda = 10)
ynegbinom1 <- dnbinom(x, size = 4, mu = 10)
ynegbinom2 <- dnbinom(x, size = 400, mu = 10)
dat1 <- data.frame(x = rep(x, times = 2),
Probability = c(ypois1, ynegbinom1, ypois1, ynegbinom2),
Distribution = rep(rep(c("Poisson","Neg. binom"),
each = length(x)), 2),
Theta = rep(c(4,400), each = 2*length(x)))
ggplot(dat1, aes(x, Probability, color = Distribution)) +
geom_point(size = 2) +
geom_line(linetype = 2) +
facet_wrap(~Theta, labeller = label_both)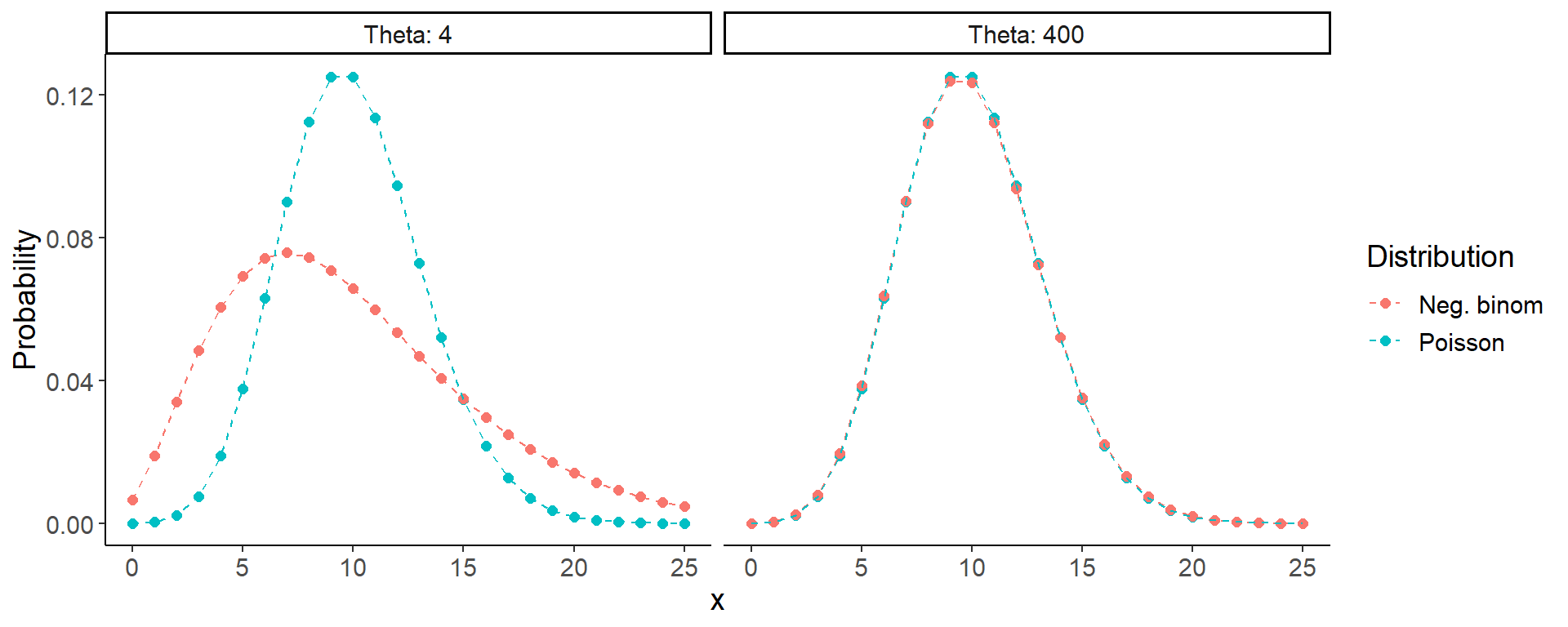
- \(\mu = 10\) in all panels
- What are the variances (according to previous slide)?
:::
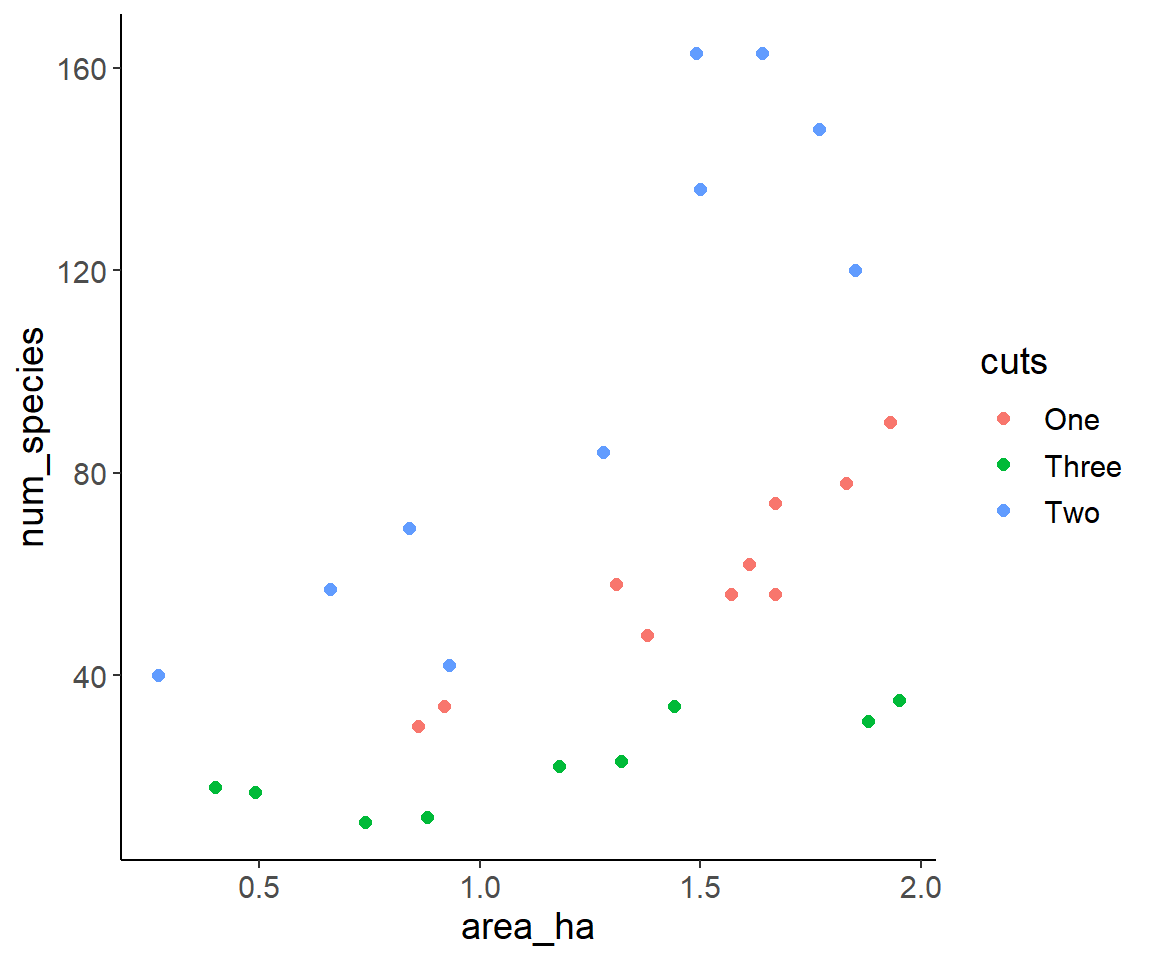
Insect diversity in urban grasslands
Inspired by FU project Flowering campus
Question: How does insect species number vary with mowing frequency and area of the grassland sites?
Data set: samples from 30 grassland sites
08_insect_diversity.csvVariables:
cuts:number of mowing events per year (consider as categorical variable!)area_ha:area of the grassland site in hectarnum_species:total number of insect species over the catching period


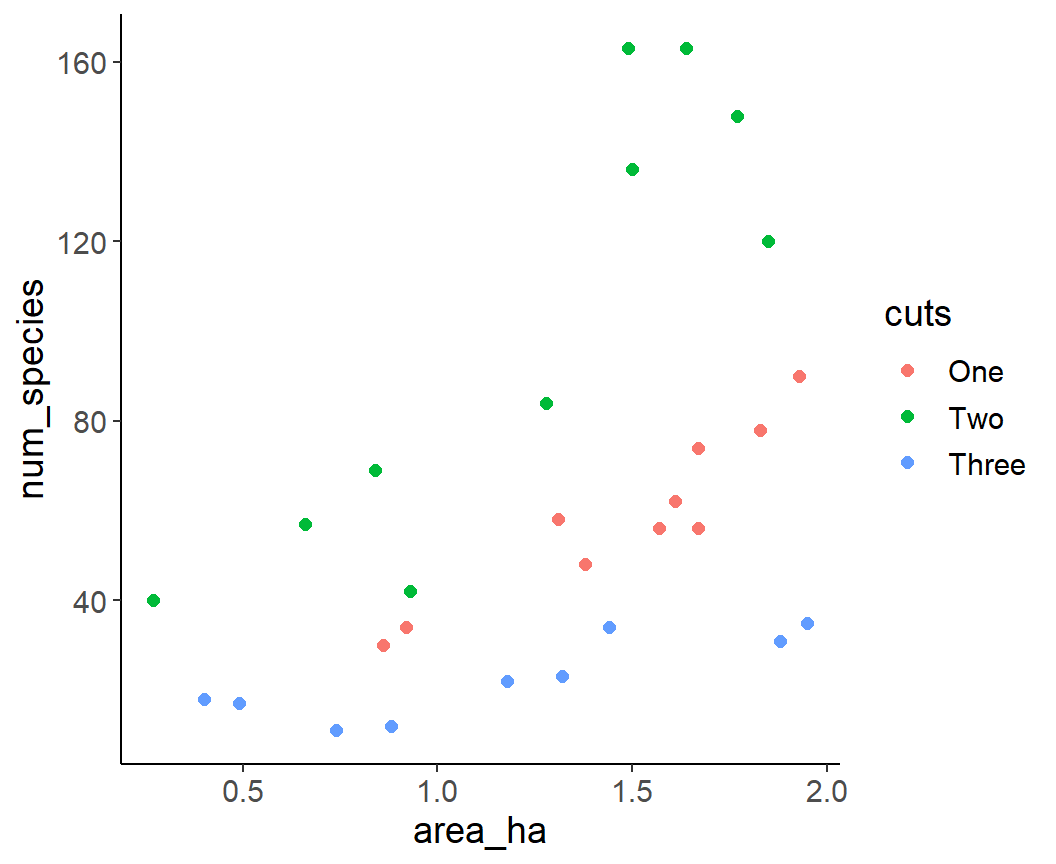
Read and plot
Adjust categorical variable
Fit GLM model
- Test interaction effect between area and no. of cuts naively.
glm1 <- glm(num_species ~ area_ha*cuts,
family = "poisson", data = insects1)
drop1(glm1, test = "Chi")Single term deletions
Model:
num_species ~ area_ha * cuts
Df Deviance AIC LRT Pr(>Chi)
<none> 66.677 249.65
area_ha:cuts 2 71.325 250.30 4.6487 0.09784 .
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1- Marginally significant interaction with p < 0.1
Check for overdispersion!
Call:
glm(formula = num_species ~ area_ha * cuts, family = "poisson",
data = insects1)
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 2.653846 0.219913 12.068 < 2e-16 ***
area_ha 0.929122 0.137494 6.758 1.4e-11 ***
cutsTwo 0.752805 0.245878 3.062 0.0022 **
cutsThree -0.276323 0.289262 -0.955 0.3394
area_ha:cutsTwo -0.009462 0.156264 -0.061 0.9517
area_ha:cutsThree -0.330170 0.191993 -1.720 0.0855 .
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for poisson family taken to be 1)
Null deviance: 862.900 on 29 degrees of freedom
Residual deviance: 66.677 on 24 degrees of freedom
AIC: 249.65
Number of Fisher Scoring iterations: 4Residual deviance / degrees of freedom = 66.7/24 = 2.77 –> Clear overdispersion!
Fit quasi-poisson model in R
Call:
glm(formula = num_species ~ area_ha * cuts, family = "quasipoisson",
data = insects1)
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 2.653846 0.362503 7.321 1.46e-07 ***
area_ha 0.929122 0.226646 4.099 0.00041 ***
cutsTwo 0.752805 0.405305 1.857 0.07557 .
cutsThree -0.276323 0.476819 -0.580 0.56764
area_ha:cutsTwo -0.009462 0.257586 -0.037 0.97100
area_ha:cutsThree -0.330170 0.316481 -1.043 0.30723
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for quasipoisson family taken to be 2.717213)
Null deviance: 862.900 on 29 degrees of freedom
Residual deviance: 66.677 on 24 degrees of freedom
AIC: NA
Number of Fisher Scoring iterations: 4Tests with Quasi-Likelihood
- Testing hypothesis in quasi-likelihood models –> F-Test
Single term deletions
Model:
num_species ~ area_ha * cuts
Df Deviance F value Pr(>F)
<none> 66.677
area_ha:cuts 2 71.325 0.8367 0.4454- Clearly not significant, after accounting for overdispersion!
Negative binomial GLM in R
- Function
glm.nb()from packageMASS
Call:
glm.nb(formula = num_species ~ area_ha * cuts, data = insects1,
init.theta = 53.69134429, link = log)
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 2.66114 0.29494 9.023 < 2e-16 ***
area_ha 0.92437 0.18910 4.888 1.02e-06 ***
cutsTwo 0.70754 0.33672 2.101 0.0356 *
cutsThree -0.27814 0.36683 -0.758 0.4483
area_ha:cutsTwo 0.02343 0.22230 0.105 0.9160
area_ha:cutsThree -0.32990 0.24785 -1.331 0.1832
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for Negative Binomial(53.6913) family taken to be 1)
Null deviance: 402.868 on 29 degrees of freedom
Residual deviance: 26.739 on 24 degrees of freedom
AIC: 232.64
Number of Fisher Scoring iterations: 1
Theta: 53.7
Std. Err.: 24.6
2 x log-likelihood: -218.643 Negative binomial GLM in R
- Testing hypothesis in negative binomial GLMs –> Likelihood Ratio Test with Chi2-distribution
Single term deletions
Model:
num_species ~ area_ha * cuts
Df Deviance AIC LRT Pr(>Chi)
<none> 26.739 230.64
area_ha:cuts 2 30.099 230.00 3.3595 0.1864Overdispersion